Advanced Practical Genetics
Microarray - Hands-on data analysis
203.305
Dr. Pierre-Yves Dupont, Postdoctoral Researcher
p.y.dupont@massey.ac.nz
Planning
25.9.17 - 9-10am - AHB1.40B
Introduction (lecture)
2.10.17 - 9-10am - AHB1.40B
Paper discussion
3.10.17 - 10-1pm - C5.15
From raw data to lists of differentially expressed genes (Step by step analysis of a microarray data set using the R language)
9.10.17 - 9-10am C5.15
Lab discussion
10.10.17 - 10-1pm C5.15
Biological interpretation of microarray data (Gene ontology analysis using the R language + online research of candidate genes)
Microarray studies
- Introduction
- Microarray technology
- Analysis
- MIAME
- Examples of microarray studies (paper discussion topic and lab topic)
What are microarrays?
A microarray is a solid support (such as a membrane or glass microscope slide) on which DNA of known sequence is deposited in a grid-like array.

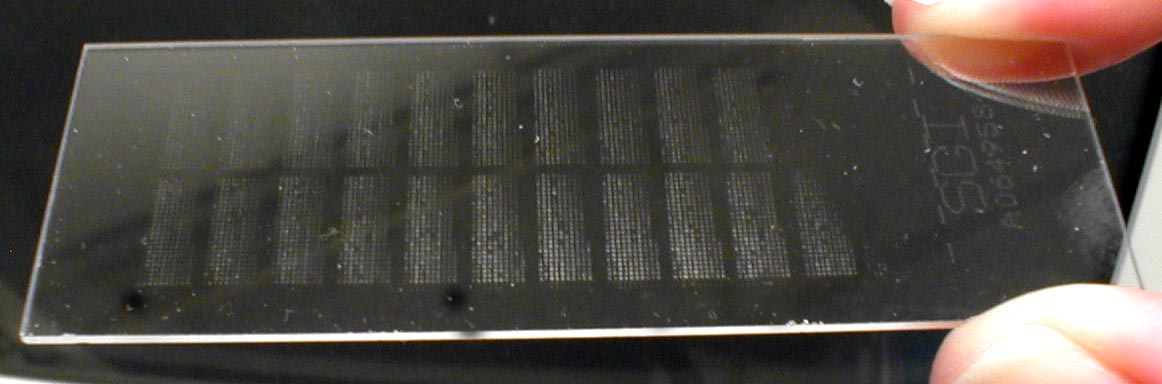
What are microarrays?
DNA microarray
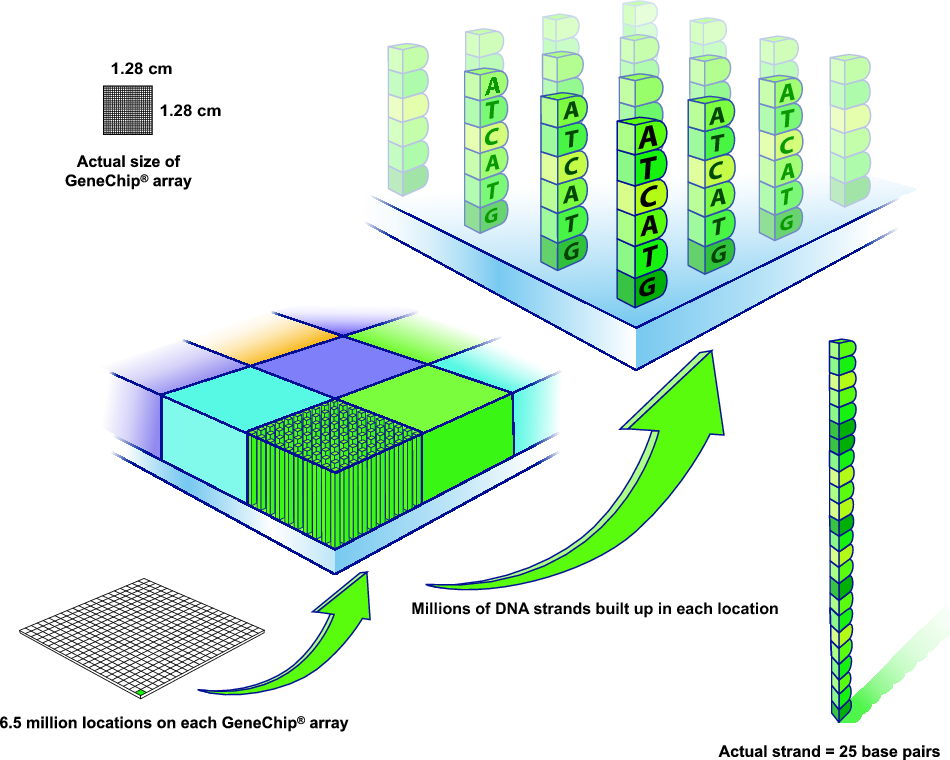
What are microarrays
Hybridisation and transcriptomics?
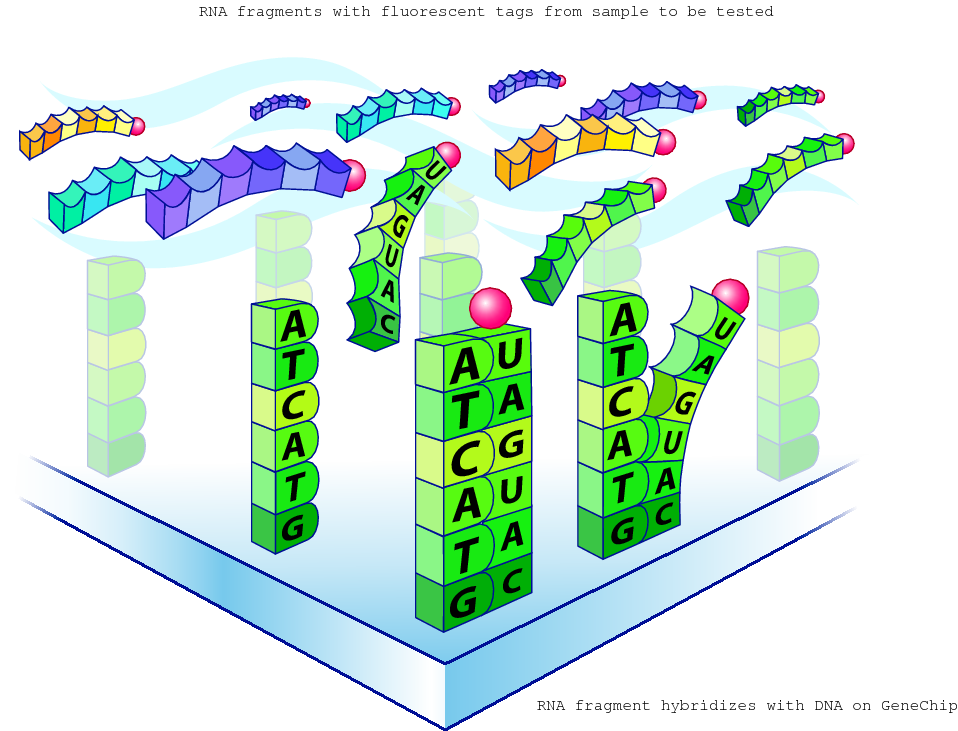
The amount of RNA hybridised on each grid location can be measured and is a proxy for the gene expression level
Microarray applications
- Gene expression analysis
- Re-sequencing
- SNP-analysis
- DNA-Protein interactions
Expression Studies
Figure modified from: Katherine Joyce, Woods Hole Oceanographic Institution
Definitions
- Genome: entire DNA sequence of an organism
- Epigenome: chemical marks of the genome that modify its expression
- Transcriptome: all gene transcripts present in a given cell/tissue at a given time (“snapshot”)
- Transcriptomics: global analysis of gene expression = genome-wide expression profiling
Definitions
- cDNA: complementary DNA made from mRNA by the enzyme reverse transcriptase
- EST: Expressed Sequence Tag, small pieces of an expressed gene (cDNA)
- Hybridisation: based on complementary molecules, sequences that are able to base-pair with one another. When two complementary sequences find each other, they will lock together, or hybridise (primer annealing, probe-target binding etc).
Genome-wide expression studies - Medical applications
- Cancer research: Cell-cycle monitoring, genetic markers detection
- Drug development and response: Treatment-induced expression pattern
- Diagnosis: Disease-associated expression patterns
Genome-wide expression studies - Biological applications
- Development biology: comparison of different developmental stages
- Ecology: interactions between organisms (symbiosis, pathogenicity...) or between organisms and environment (temperature, nutrient...)
- Evolution: within and between species variation, hybrids vs. parents, diploids vs. polyploids
- Functional analyses: wild type vs. mutant
Microarray studies
- Introduction
- Microarray technology
- Analysis
- MIAME
- Examples of microarray studies (paper discussion topic and lab topic)
Microarray analysis principle
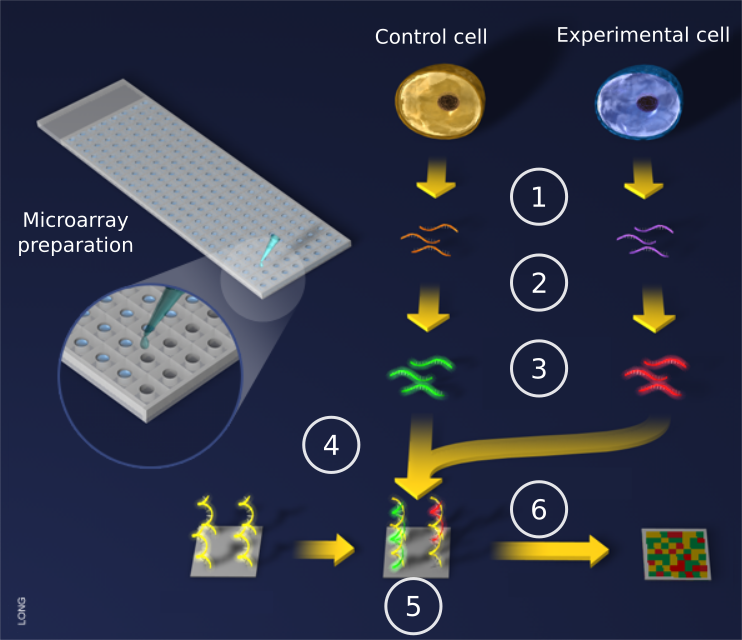
Image from: http://www.scq.ubc.ca/image-bank/
Microarray analysis principle

Image from: http://www.scq.ubc.ca/image-bank/
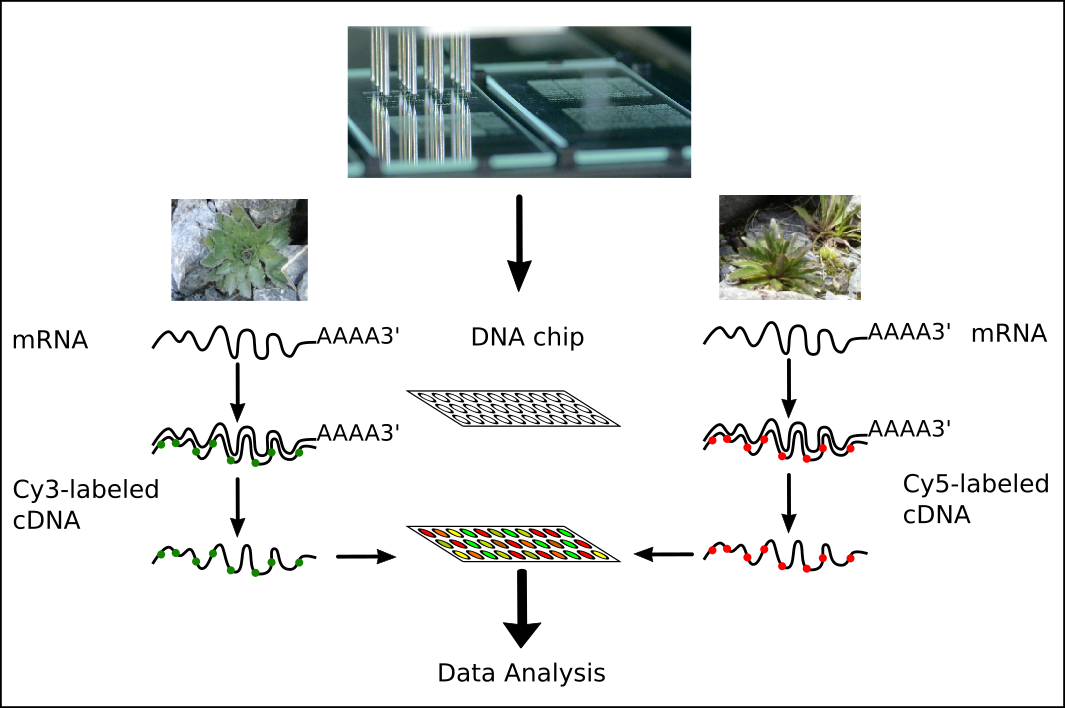
Competitive hybridisation
It is possible to represent different samples on one microarray using different fluorescent molecules (fluorophores)
- Cyanin 3 (Cy3): green fluorescence (excited at 550nm, emission at 570nm)
- Cyanin 5 (Cy5): red fluorescence (excited at 650nm, emission at 770nm)
Competitive hybridisation
Competitive hybridisation
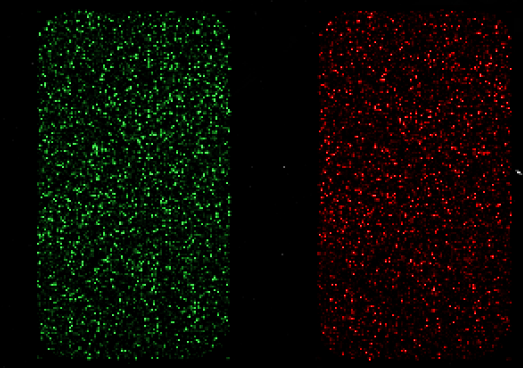
Competitive hybridisation
Microarray study pipeline
Question driven
Goals? Hypothesis? Questions?
Microarray study pipeline
- Platform/ design
- What technology?
- Source of the gene probes?
- Cross-species hybridisation?
- Replication level
- Hybridisation scheme
Microarray study pipeline
- Platform
- Laboratory steps
- Sample preparation and labelling
- Hybridisation
- Washing
- Image acquisition
Microarray study pipeline
- Platform
- Laboratory steps
- Bioinformatics steps
- Data transformation and normalisation
- Analysis of differentially expressed genes (statistical tests, gene ontology, ...)
- Visualisation (graphics)
- Data storage (databases, MIAME standards)
Microarray study pipeline
- Platform
- Laboratory steps
- Bioinformatics steps
- Data interpretation
- Answers?
- New hypotheses?
- Follow-up experiments?
- Validation?
Microarray studies
- Introduction
- Microarray technology
- Analysis
- MIAME
- Examples of microarray studies (paper discussion topic and lab topic)
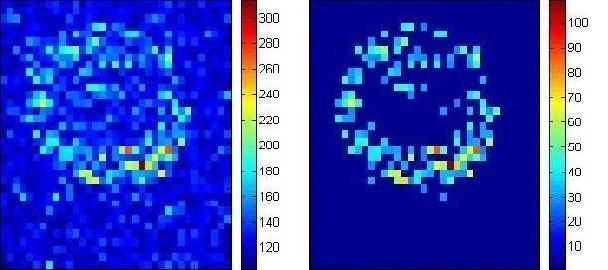
Noise reduction
Before
After
Data normalisation
Global normalisation
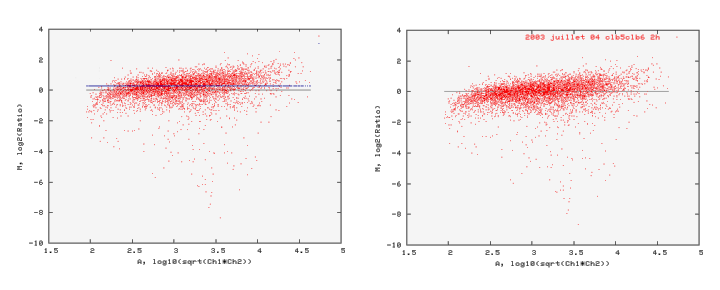
Data normalisation
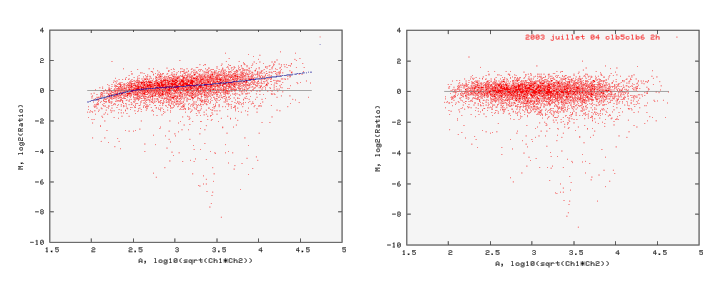
Lowess normalisation (LOcally WEighted Scatterplot Smoothing)
Expression ratios, M & A
- $\color{green}{Cy3} = Sample1$ (Green)
- $\color{red}{Cy5} = Sample2$ (Red)
- $\color{red}{Cy5}$ > $\color{green}{Cy3}$: higher expression in sample 2
- $\color{green}{Cy3}$ > $\color{red}{Cy5}$: higher expression in sample 1
- Log fold ratio: $M = log_2({\color{red}{Cy5} \over \color{green}{Cy3}})$ = $log_2({\color{red}{Cy5}}) - log_2({\color{green}{Cy3}})$
- Expression average: $A = {1\over2} (log_2(\color{red}{Cy5}) + log_2(\color{green}{Cy3}))$ = ${1\over2}log_2(\color{red}{Cy5} \color{green}{Cy3})$
Log Fold Ratio
Expression ratio: $log({\color{red}{Cy5} \over \color{green}{Cy3}})$
Log Fold Ratio
Reminder: $log_2(x)$ is the unique real number $y$ such that: $2^y = x$.
For example: $log_2(8) = 3$ because $2^3 = 8$
| $\color{red}{Cy5}/\color{green}{Cy3}$ | $log_2({\color{red}{Cy5}/\color{green}{Cy3}})$ |
|---|---|
| 4 | 2 |
| 2 | 1 |
| 1 | 0 |
| 0.5 | -1 |
| 0.25 | -2 |
Hypothesis testing
T-test
Null hypothesis ($H_0$): gene $x$ is not differentially expressed between two treatments
Mean:
$\overline{x}={{1 \over M} \sum_{i=1}^{M}{x_i}}$; for gene $x$ in M replicates
Variance:
$S_{x}^2 = {{1 \over {M-1}} \sum_{i=1}^{M}({x_i^2 - \overline{x}^2})}$
T-statistic:
$T_x={{ \overline{x_{C_1}} - \overline{x_{C_2}} } \over \sqrt[2]{ {S_{x_{C_1}}^2 \over M} + {S_{x_{C_2}}^2 \over N} } }$
T-test and P-value
T-test is used only to compare two samples. To compare more than two samples, ANOVA (ANalysis Of VAriance) is used.
T-test and P-value
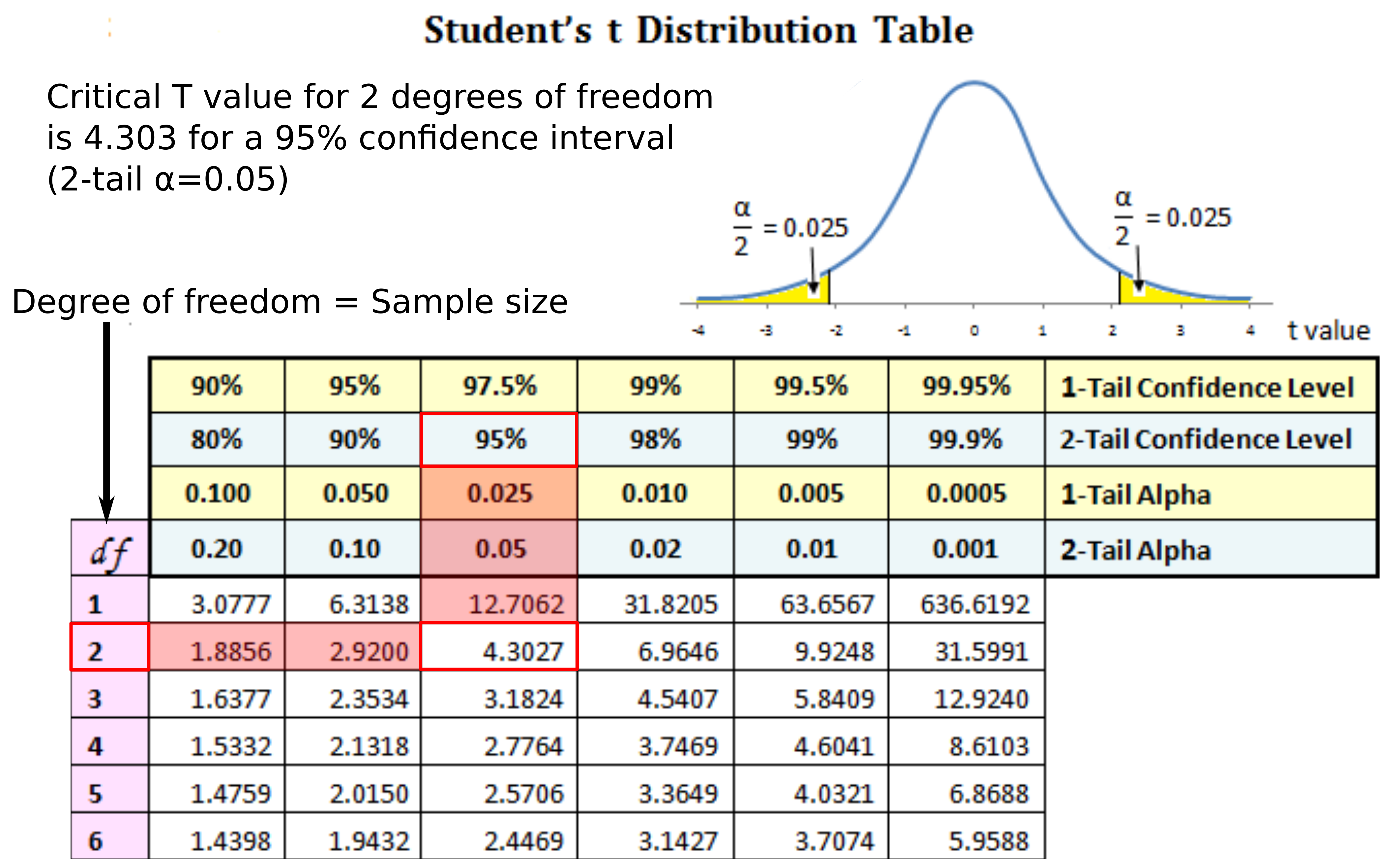
Hypothesis testing
T-test
Null hypothesis ($H_0$): gene A is not differentially expressed between two treatments
- Compute the signal to noise ratio (difference of the means or medians) for each gene
- Compute the t-statistic for each gene using the replicates
- Compare t-statistic with the t-distribution
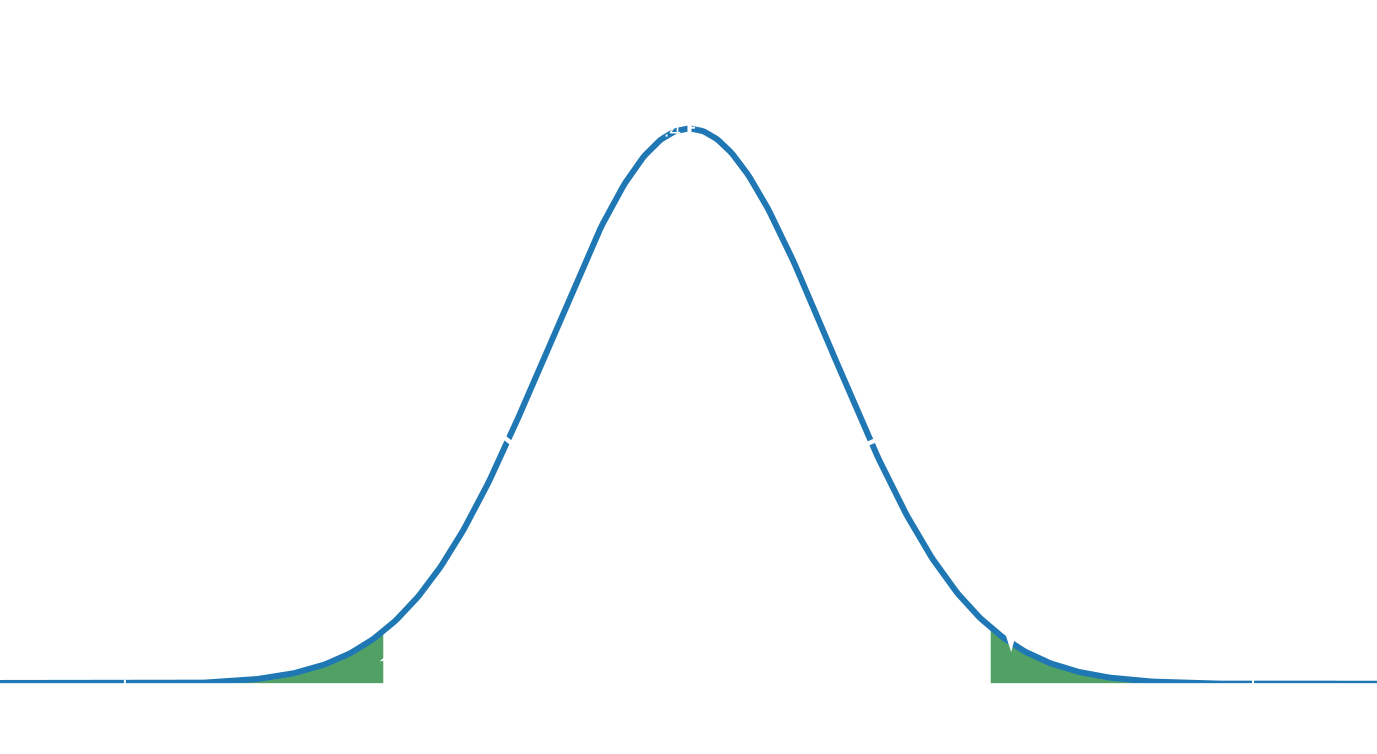
- If t-statistic is more extreme than the critical t-statistic at a chosen significance level (e.g. $\alpha=0.05$) reject the null hypothesis, otherwise accept it. P-value estimation
Quiz
Usually, a p < 0.05 is considered small enough to reject the null hypothesis of no biological effect in favour of the alternative hypothesis of a biological effect.
P-values are also known under type 1 error – the probability of rejecting the null hypothesis when it is actually true (= false positive rate).
P-value of 0.01 means a false positive rate of 1 %.
When analysing multidimensional data sets, p-values need to be adjusted for multiple testing .
Two common p-value adjustment methods are Bonferroni and False Discovery Rate .
Bonferroni Correction
- If you hypothesize that a specific gene is up-regulated, p < 0.05 is fine.
- If you hypothesize that any of 10,000 genes is up-regulated, with p < 0.05 you can expect to see 5% (500 genes) up-regulated by chance alone.
- To account for the thousands of repeated measurements, some researchers apply a Bonferroni correction.
$p$ < $(0.05)/10,000$
$p$ < $5e^{-6}$
$p$ < $5e^{-6}$
The Bonferroni correction is generally considered to be too conservative and False Discovery Rate (FDR) should be used.
False Discovery Rate
Benjamini-Hochberg method
Imagine an array with 6400 genes and an experiment where 184 genes are differentially expressed at $P=0.01$: 64 genes would be expected to appear differentially expressed by chance alone.
FDR = false discovery rate = ${{64}\over{184}} * 100 = 35 \%$
False Discovery Rate
Benjamini-Hochberg method
| P-value | Observed number of genes | Expected number of false positives | FDR |
|---|---|---|---|
| $10^{-2}$ | 184 | 64 | 35 |
| $10^{-3}$ | 35 | 6 | 18 |
| $10^{-4}$ | 15 | 0.6 | 4 |
With decreasing p-value, FDR also decreases, but so does the number of differentially expressed genes – choose a p-value which balances both!
Microarray studies
- Introduction
- Microarray technology
- Analysis
- MIAME
- Examples of microarray studies (paper discussion topic and lab topic)
MIAME Standard
Minimum Information About a Microarray Experiment that is needed to enable the interpretation of the results of the experiment unambiguously and potentially to reproduce the experiment
MIAME Standard
- Raw data for each hybridisation (CEL or GPR files)
- Processed (normalised) data (used to draw the conclusions from the study)
- Essential sample annotation including experimental factors and their values
- Experimental design including sample data relationships (e.g. which hybridisations are technical and biological replicates)
- Sufficient array annotation (e.g. gene identifiers, probe sequences)
- Essential laboratory and data processing protocols (e.g. normalisation method used to obtain the final data)
Gene expression databases
Gene Expression Omnibus (GEO) @NCBI (http://www.ncbi.nlm.nih.gov/geo/)
![]()
Gene expression databases
Geo Datasets @NCBI (http://www.ncbi.nlm.nih.gov/gds/)
Geo Profiles @NCBI (http://www.ncbi.nlm.nih.gov/geoprofiles/)![]()
Geo Profiles @NCBI (http://www.ncbi.nlm.nih.gov/geoprofiles/)
Gene expression databases
ArrayExpress @ EBI (http://www.ebi.ac.uk/arrayexpress/)
![]()
Gene expression databases
Expression Atlas @ EBI (http://www.ebi.ac.uk/gxa/)
![]()
Microarray studies
- Introduction
- Microarray technology
- Analysis
- MIAME
- Examples of microarray studies (paper discussion topic and lab topic)
Microarray paper discussion
Lab case study: Herbivory in Nicotiana attenuata (Solanaceae)

|
What type of research?
|
Lab case study: Herbivory in Nicotiana attenuata (Solanaceae)

|
Why N. attenuata?
|
Case study - Chips, veggies & vegetarians

Case study - Chips, veggies & vegetarians
| The chip: cDNA array with 15,264 potato genes from TIGR (The Institute for Genomic Research) |  |
||||
| The veggies | The vegetarian | ||||
| Solanum nigrum Black nightshade |
 |
Nicotiana attenuata Coyote tobacco |
 |
Manduca sexta |  |
- Question:
- Do tobacco and black nightshade plants respond differently to caterpillar attack?
Microarray Case Study
RNA source
2 herbivore treatments and 2 controls
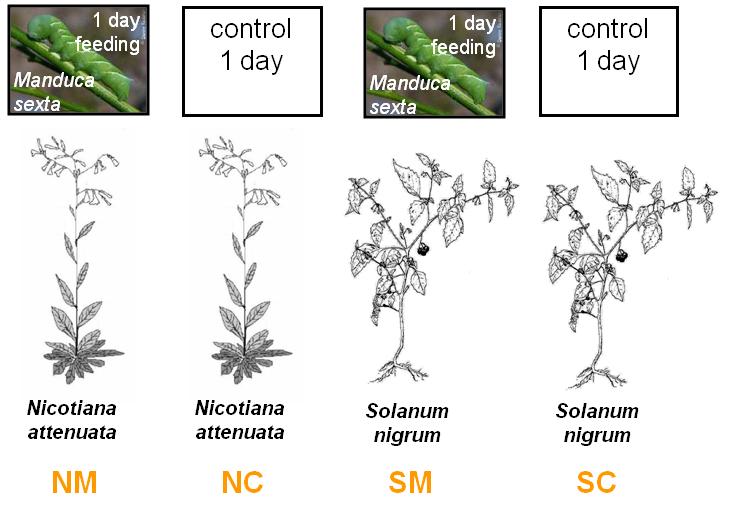
Microarray Case Study
6 arrays
Each arrow represents one array. Herbivore-induced tissue (Cy3) was co-hybridised control tissue (Cy5). Each comparison was replicated three times.
What will you do in the lab?
- Lab 1
- R warm-up exercise. Identification of differentially expressed genes
- Lab 2
- Identification of differentially expressed biological processes